Data integration for inferring biogeographic history
The ability of species to move to suitable regions is critical to their capacity to cope with climate change. Understanding species’ responses to past environmental changes is needed to forecast their responses to ongoing climate change. Our lab is developing methods for integrative post-glacial tree range dynamics modeling. This research project is a collaboration with John Robinson (Michigan State University), Andria Dawson (Mount Royal University), Sean Hoban (The Morton Arboretum), Adam Smith (Missouri Botanical Garden), and Allan Strand (College of Charleston).

Our TIMBER group. On the top, from left to right, Dr. Andria Dawson (Mount Royal University), Dr. John Robinson (Michigan State University), Dr. Allan Strand (College of Charleston), Lauren Jenkins (Duke University), Dr. Adam Smith (Missouri Botanical Garden). A the bottom, Dr. Sean Hoban (The Morton Arboretum), Dr. Antonio R. Castilla (Oklahoma State University) and our team friend Lainey.
Our research integrates fossil pollen, occurrence data via species distribution models, and population genomics to quantify historical range shifts. Using Approximate Bayesian Computation, we are developing an integrative analytical framework to combine these three sources of information to infer demographic parameters, the location of refugia, and the pace of range movement (more information in Hoban et al. 2019). We are implementing our integrative approach in the R package holoSimCell.
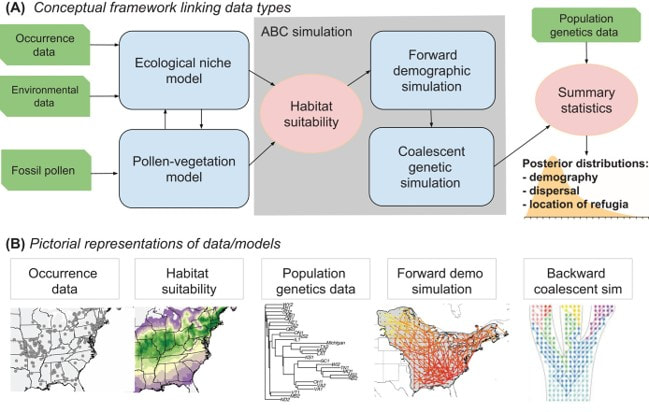
Conceptual diagram showing our framework to integrate multiple data types via Approximate Bayesian Computation. Modified from Hoban et al. (2019): Hoban, S., Dawson, A., Robinson, J. D., Smith, A. B., & Strand, A. E. (2019). Inference of biogeographic history by formally integrating distinct lines of evidence: genetic, environmental niche and fossil. Ecography 42: 1991-2011.
Adaptive introgression in the oak syngameon
The rapid creation of novel allelic combinations may be critical in rapidly changing environments, like those predicted under climate change, when standing genetic variation may only offer limited adaptive potential and dispersal to environmentally suitable regions is limited by human landscape alteration. Adaptive introgression can result in favorable allelic combinations within the new genomic background and accelerate adaptation. Our lab is currently addressing this topic in the syngameon formed by shinnery oak (Quercus havardii) and the hybridizing species in southern North America. We are using an integrative approach that combines genomic and experimental work to investigate whether shinnery oak has adapted to sub-optimal environments through introgressed alleles of congeneric species.
Drivers of gene flow in oaks of the Chihuahuan desert region
At Castilla lab, we are using landscape genomic tools to investigate the primary drivers of gene flow in tree populations of desertic regions. Specifically, we are working with gray oak (Quercus grisea Liebmann), a tree species with a highly fragmented distribution that typically occurs on ridges, rocky slopes, and the banks of streams. We are using genomic data and GIS tools to investigate the primary landscape features affecting the genetic connectivity of gray oak populations in the Chihuahuan desert.
Gray oaks (Q. grisea) in the Chihuahuan desert showing their characteristic association with banks of streams at the bottom of canyons. (Photo credit: Castilla Lab)
Previous studies widely reported hybridization between gray oak and Gambel oak (Q. gambelii Nutt) western wards. Towards the eastern portion of its range, gray oak and sandpaper oak (Q. pungens Liebm.) are sympatric, and their populations present abundant individuals with intermediate phenotypic traits. Our lab is examining a possible hybridization between gray oak and sandpaper oak which could make gray oak a good candidate for a conduit species within the oak syngameon. To address this goal, we are collaborating with plant taxonomists to incorporate morphological and genomic information into the delineation of species.
Differences in leaf morphology between a gray oak individual (on the left) and a possible hybrid between gray oak and sandpaper oak (on the right). (Photo credit: Castilla Lab)








